Welcome
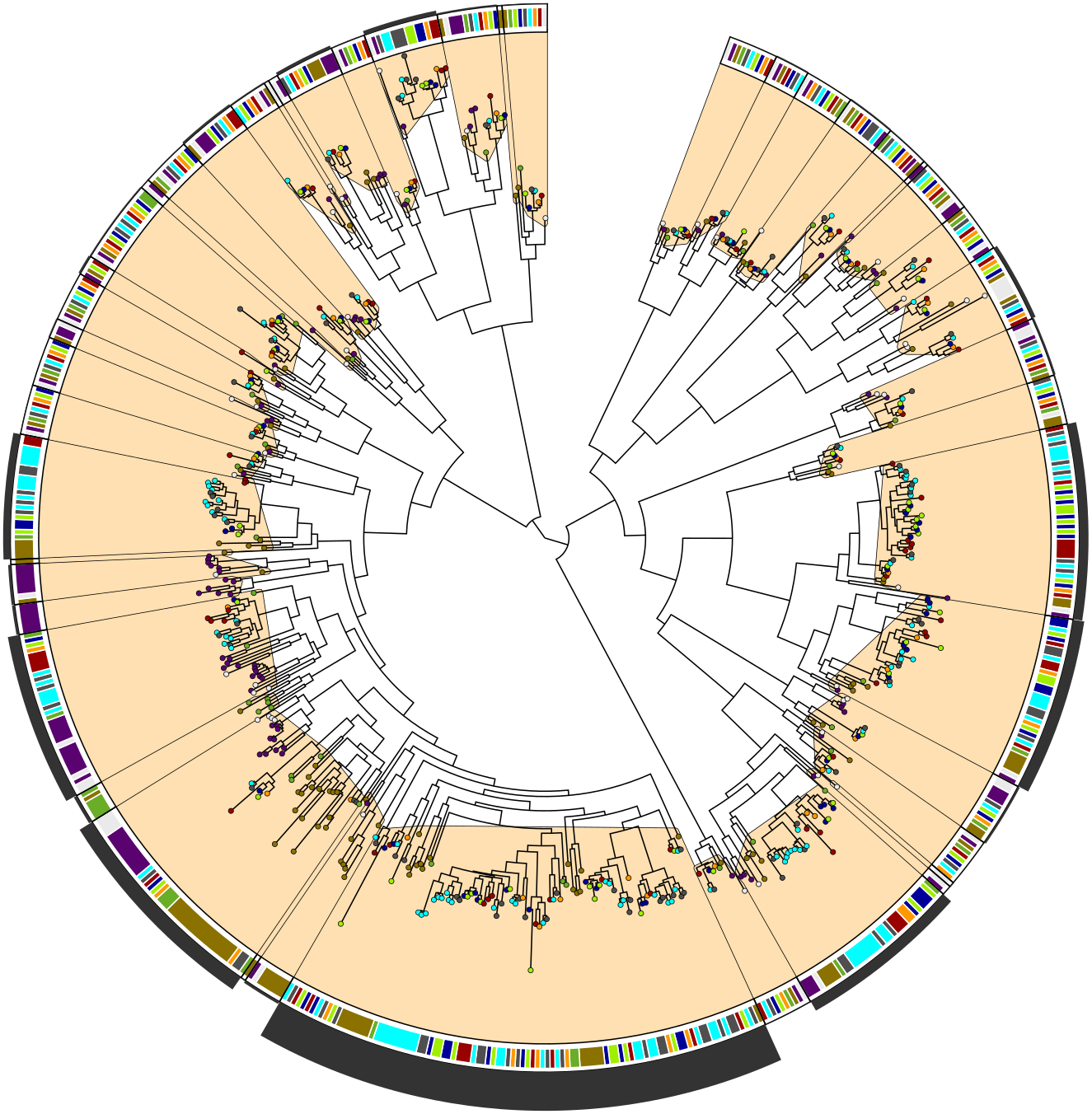
Welcome to MIPhy, a tool to quantify phylogenetic instability in trees of multi-species gene families. See the Documentation tab above for more details and for a tutorial on how to use MIPhy. It can also be downloaded as a stand-alone program from GitHub.
MIPhy was created to identify rapidly evolving genes quickly and easily for you. It is a kind of meta-phylogenetic analysis, as it requires that you perform a phylogenetic analysis, and then it analyzes those results. In your phylogenetic tree, a "phylogenetically stable" group of sequences would contain exactly one from each species under consideration, and the relationships between those sequences would exactly mirror the relationships between those species. MIPhy clusters your tree into minimum instability groups (MIGs), and assigns each MIG an instability score quantifying how much it deviates from that expected pattern.
MIPhy inputs
Upload summary
No files uploaded.
Note: the parameter weights are set once you press the upload button.